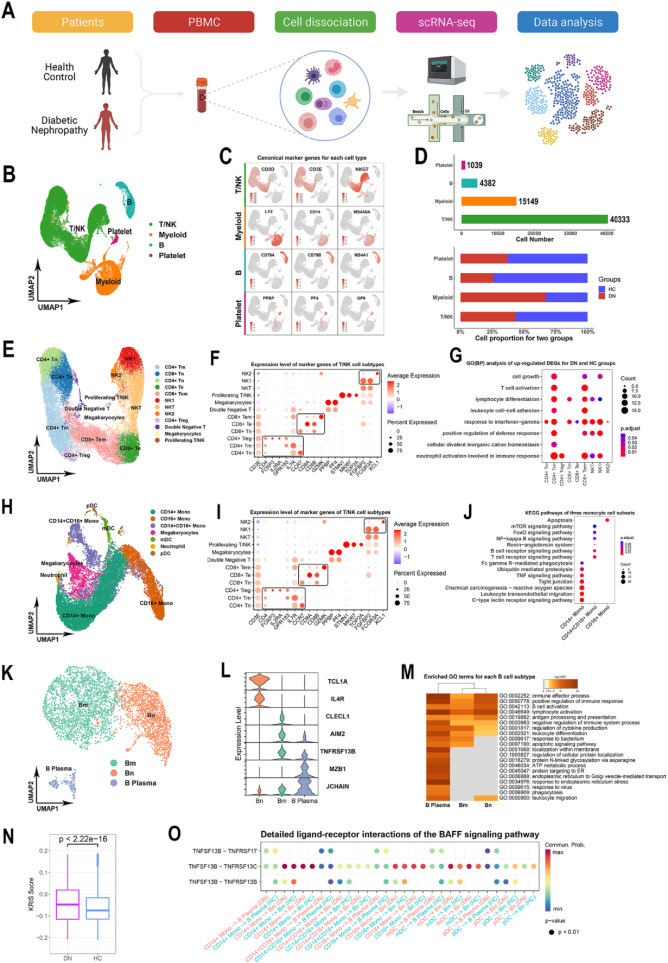
Single-cell transcriptomics systematically discloses the immune-inflammatory responses of peripheral blood mononuclear cells for diabetic nephropathy


Diabetic nephropathy (DN) has become the leading cause of end-stage renal disease with high morbidity and mortality among individuals with diabetes mellitus. Although functional alterations of renal infiltrating immune cells have been reported as part of the pathological mechanism of DN, the understanding of the immune response underlying peripheral blood mononuclear cells (PBMCs) in DN remains limited. Here, single-cell RNA sequencing (scRNA-seq) was used to profile the transcriptomic signatures of PBMCs from DN patients. We identified four well-known cell types of PBMCs and analyzed their respective cell subtypes. The underlying biological processes were captured from the altered cell (sub)types. A number of cell-type-specific cytokines and transcription factors driving the DN-associated transcriptomic changes were detected. Taken together, our data revealed detailed and rich single-cell transcriptomic signatures of PBMCs from DN patients and identified cell-type-specific pathways and molecules associated with the mechanisms of DN progression.