Incidental and secondary findings in trio exome sequencing


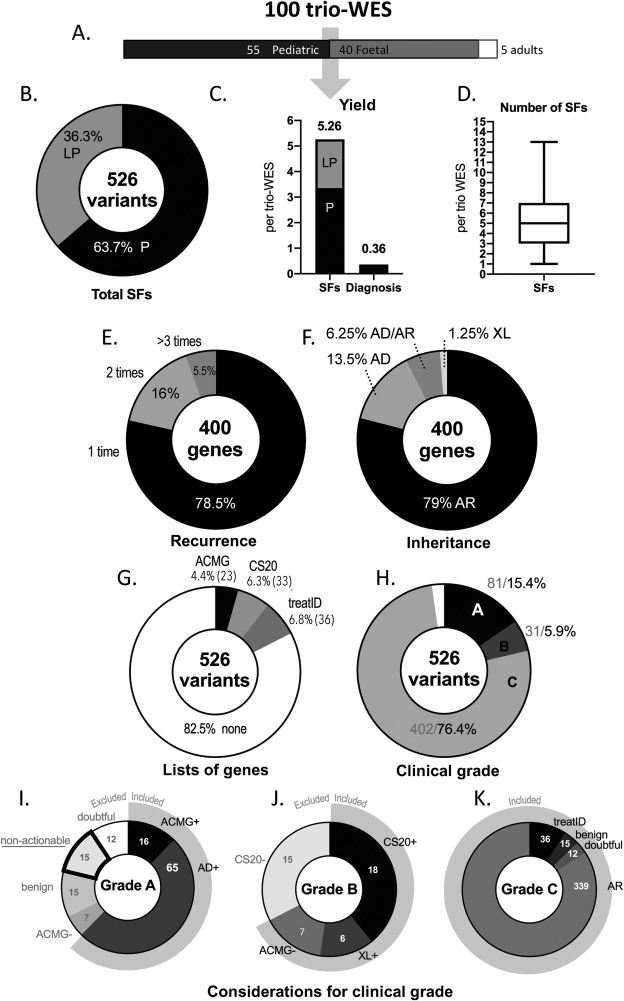
Exome sequencing (ES) generates secondary findings (SFs) in 2 % of tested individuals if one follows the American College of Medical Genetics and Genomics (ACMG) guidelines.1,2 However, the rate of incidental and secondary findings (ISFs) is higher in routine clinical practice because of (i) the use of trio ES instead of solo sequencing and (ii) the exclusion of the incidental findings (IFs) of medical value concerning genes in the ACMG list. Hence, it is not clear how sufficient is a restricted list of genes to detect every ISF of major clinical value; and what is the amount of additional workload for the laboratory. Using 100 trio ES datasets, we determined the proportion of pathogenic or likely-pathogenic ISFs and their clinical value (See supplementary file 1 for Materials and Methods). We evaluated the accuracy of three lists of genes (see supplementary file 2) for detection of SFs: the 2021 ACMG v3 SFs list, a list of genes involved in treatable intellectual disabilities3 (treat-ID list), and a list of genes involved in the 20 most frequent diseases in general populations (CS20 list).